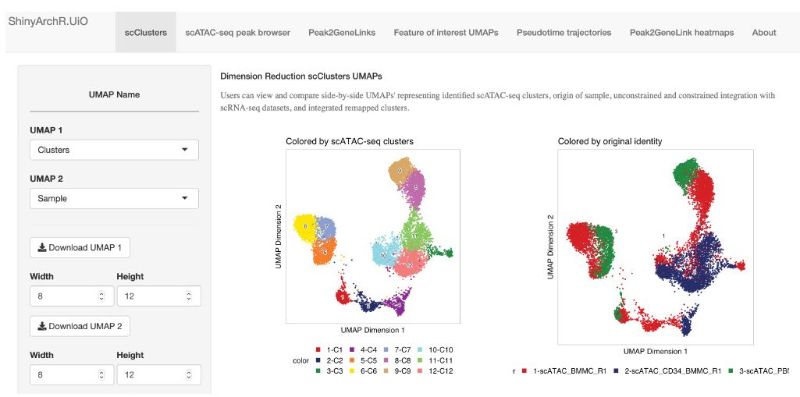
ShinyArchR.UiO is a tool that can streamline collaborative efforts for interpretation of large chromatin accessibility datasets and promote open access data sharing for wider audiences. Try a demo version of ShinyArchR.UiO here. This is a project mainly under the GPL license version 3 and the code is open, please go to EskelandLab Github.
Sharma, A., Akshay, A., Rogne, M., and Eskeland, R. (2021). ShinyArchR.UiO: user-friendly, integrative and open-source tool for visualization of single-cell ATAC-seq data using ArchR. Bioinformatics.
Learn more about ShinyArchR.UiO in our short Introduction video.