What is your field of research?
Our Computational Biology & Gene Regulation group is working in the field of Computational Biology; sometimes referred to as Bioinformatics.

In a nutshell, we develop and assess computational methods to analyze genomic sequences (DNA). A computer scientist by background, I have gained a strong understanding of the underlying biology, and I combine this knowledge with my computational skills to develop software useful for the research community.
The goal of my group is to create the next generation of cutting-edge algorithms and open computational biology software, with immediate application to real-life biological problems.
Specifically, our computational biology research program aims at furthering the understanding of gene expression regulation (when and where genes are expressed), and the mechanisms by which it can be disrupted in human diseases such as cancer.
What are you working on at the moment and what do you hope to find out with your research?
- In the context of personalized medicine, it is becoming critical to decipher how mutations specific to an individual can trigger a disease. Thanks to high-throughput sequencing technologies (also known as next-generation sequencing technologies), we have unprecedented opportunities to study the human genome in the context of diseases. While most studies have focused on genomic regions encoding for proteins (and representing only ~2% of the human genome), we try to predict which mutations in cis-regulatory DNA regions (switches to regulate when and where genes are transcribed from DNA to RNA) are causal for diseases.
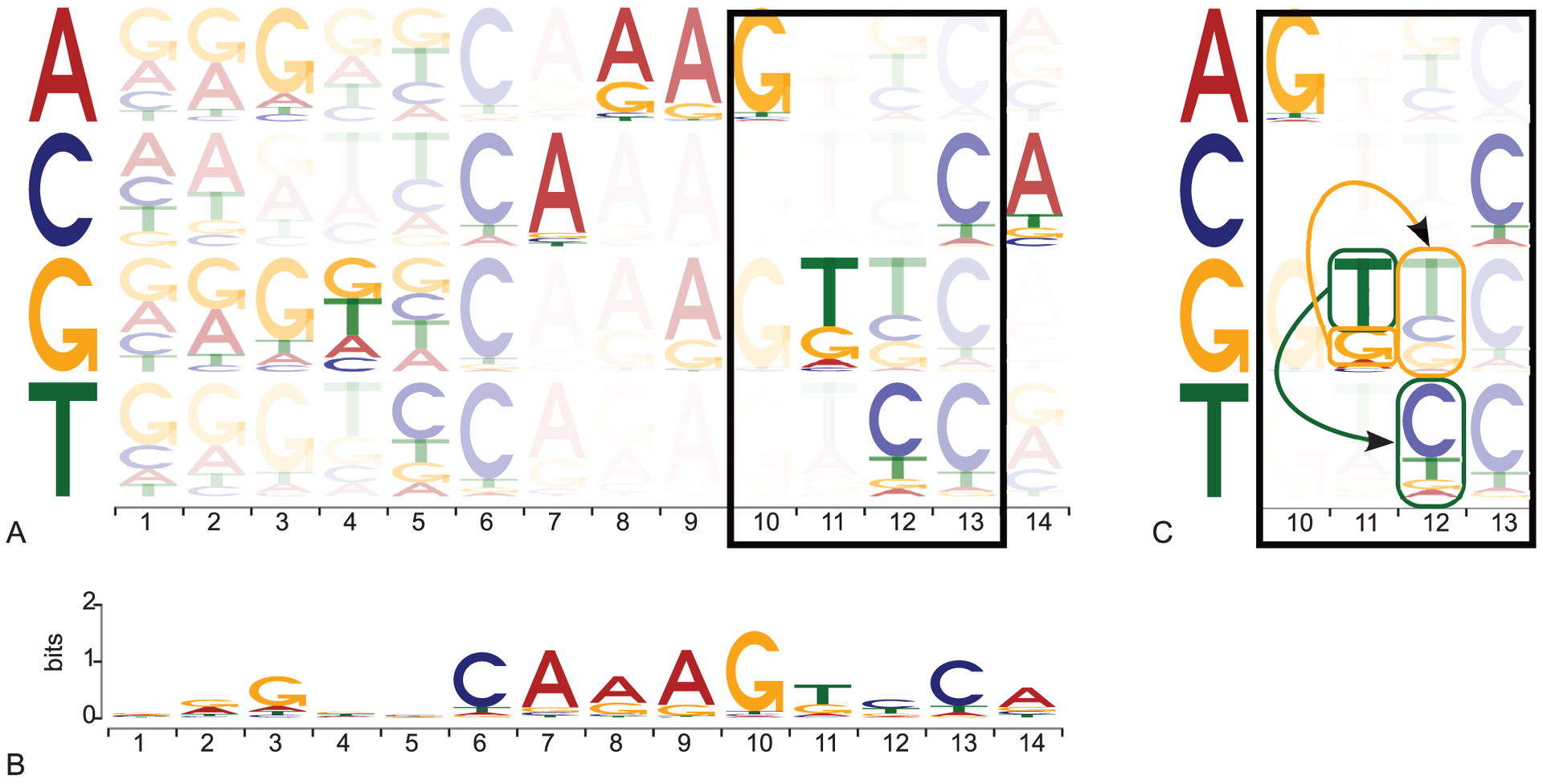
Transcription factors (TFs) are key proteins binding to these switches to control when, where, and to what extent genes are transcribed. It has been shown that mutations within TF binding sites can alter gene expression, triggering human diseases such as cancer. As successful computational biology research relies on high quality data for which we have a strong understanding, we are currently combining large amounts of experimental data with in house computational models to identify the binding sites of TFs. This work will provide us with a critical map of where TFs bind in the human genome for further studies.
In terms of our next steps, we will combine whole genome sequencing and gene expression data from cancer patient samples with our high-quality regulatory map. This will provide new insights into the predictions of the impact of the mutations dysregulating gene expression and contributing to cancer.
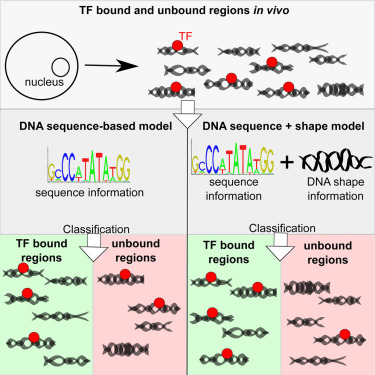
DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. Figure from A. Mathelier, B. Xin, T-P. Chiu, L. Yang, R. Rohs, W. W. Wasserman, Cell Systems 2016
Where do you think your field of research will be in ten years?
- I remember that a bioinformatics department didn’t even exist at my university (Paris 6, France’s largest university) when I started my PhD about 10 years ago. Now, it is hard to find a biology-related center or department without any bioinformatics. It shows how fast the field has grown over the years but I believe that it is still in its early stages.
With the large amount of biological data generated by individual labs and large consortia, there is a strong need for computational tools and analyses; this will only increase over the years with decreasing costs due to improved technology. In the coming decade, I believe there will be the need for a bioinformatician in every biological laboratory.
One of the most exciting things about computational biology is that it is, by essence, an interdisciplinary field. The bioinformatics, mathematics, computer science, biostatistics, and life science communities are already strongly interacting, and they will do so even more intensively in the coming years for the interpretation of biological data with direct impact on patients’ lives. Bioinformatics tools will also become key for the success of personalized medicine, where doctors and clinicians will rely on these tools to provide patients with optimal therapies.
Tell us a bit about what you were doing before you joined NCMM
- After my Master in Computer Science, I started a PhD under the supervision of Dr. Alessandra Carbone in 2006 at the Paris 6 University, France. The main focus of my research was to develop computational tools to predict short sequences of RNA (microRNAs) involved in the post-transcriptional regulation of gene expression, i.e. regulating the production of proteins from RNAs. I moved to Vancouver, Canada, in 2011 to start my postdoc with Dr. Wyeth Wasserman at the University of British Columbia. For over five years, I developed computational models to predict where transcription factors bind onto the DNA to regulate the transcription of DNA into RNA. Basically, I spent the last decade developing computational models and tools to study how gene expression is regulated.
What would you be doing if you weren’t working in research?
- It is really hard to know what I would be doing if not research. I started university thinking about becoming a math teacher, then I discovered computer science and became very interested in it and, finally, I found computational biology. The interdisciplinarity of computational biology combined with the international environment of academic science make our job extremely exciting, so I feel lucky to work in this field.
What, in your opinion, has been your greatest moment in your career so far?
- The publication of my very first paper was a great moment as it was the first big achievement. Also, I can say that I am proud of being able to launch my own laboratory here at NCMM. It feels good to work with, and mentor, talented trainees striving to learn, and to help with advancing scientific knowledge.
Tell us a bit more about your new seminar series
- We are launching the Sven Furberg Seminars in Bioinformatics and Statistical Genomics in Oslo. It is a joint effort between Geir Kjetil Sandve and Torbjørn Rognes at the Biomedical Informatics Group, Arnoldo Frigessi at the Oslo Centre for Biostatistics and Epidemiology, and myself at NCMM. Starting from March 16th, we will have monthly seminars with renowned international guest scientists who will present their computational and statistical research applied to molecular biology.
There are unparalleled opportunities arising from attending a regular seminar series with highly relevant speakers, with the added benefit of interacting with them. We expect the audience to be composed of local computer scientists, statisticians, mathematicians, and biologists interested in the development and application of computational and statistical methods and tools to analyze molecular data. The idea of these joint bioinformatics-biostatistics events is to promote scientific excellence and trigger local and international collaborations.
